10b. The Greshgorin circle theorem
[1]:
# Colab setup ------------------
import os, sys, subprocess
if "google.colab" in sys.modules:
cmd = "pip install --upgrade watermark"
process = subprocess.Popen(cmd.split(), stdout=subprocess.PIPE, stderr=subprocess.PIPE)
stdout, stderr = process.communicate()
# ------------------------------
import itertools
import tqdm
import numpy as np
import scipy.optimize
import colorcet
import bokeh.io
import bokeh.plotting
bokeh.io.output_notebook()
In the previous technical appendix, we performed a linear stability analysis on the simplest version (\(N = 1\)) of the circuit shown below.
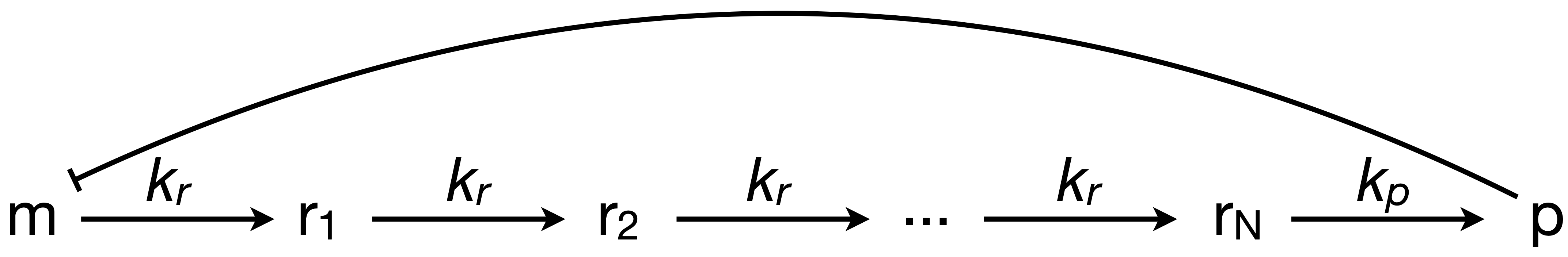
Here, we will perform a similar analysis for arbitrary \(N\) and in so doing introduce the useful Gershgorin circle theorem.
Linear stability matrix
In Chapter 10, we worked out that the dimensionless dynamical equations for this circuit are
\begin{align} &\frac{\mathrm{d}m}{\mathrm{d}t} = \frac{\beta}{1 + p^n} - m,\\[1em] \kappa^{-1}\,&\frac{\mathrm{d}r_1}{\mathrm{d}t} = m - r_i,\\[1em] \kappa^{-1}\,&\frac{\mathrm{d}r_i}{\mathrm{d}t} = r_{i-1} - r_i\; \text{for } 2 \le i \le N,\\[1em] \gamma^{-1}\,&\frac{\mathrm{d}p}{\mathrm{d}t} = r_N - p. \end{align}
There is a single fixed point \((m_0, r_{1,0}, \ldots, r_{N,0}, p_0)\),
\begin{align} m_0 = r_{1,0} = \cdots = r_{N,0} = p_0, \end{align}
with
\begin{align} \beta = m_0(1+m_0^n). \end{align}
The linear stability matrix for this system is
\begin{align} \mathsf{A} = \begin{pmatrix} -1 & 0 & 0 & \cdots & 0 & 0 & -b \\ \kappa & -\kappa & 0 & \cdots & 0 & 0 & 0 \\ 0 & \kappa & -\kappa & \cdots & 0 & 0 & 0 \\ \vdots & \vdots & \ddots & \ddots & \vdots & \vdots & \vdots \\ 0 & 0 & 0 & \ddots & -\kappa & 0 & 0 \\ 0 & 0 & 0 & \cdots & \kappa & -\kappa & 0 \\ 0 & 0 & 0 & \cdots & 0 & \gamma & -\gamma \end{pmatrix}, \end{align}
where
\begin{align} b = \frac{\beta n m_0^{n-1}}{(1+m_0^n)^2} = \frac{n m_0^n}{1+m_0^n}. \end{align}
For clarity, \(\mathsf{A}\) is an \((N+2) \times (N+2)\) matrix where the main diagonal is \((-1, -\kappa, -\kappa, \ldots, -\kappa, -\gamma)\), and the subdiagonal is \((\kappa, \kappa, \ldots, \kappa, \gamma)\). The only other nonzero entry is \(A_{1, N+2} = -b\).
Gershgorin circle theorem
We can find the eigenvalues of this matrix numerically for given values of \(\beta\), \(\gamma\), \(\kappa\), and \(n\), but we can make some analytical progress by using the Gershgorin circle theorem. The theorem goes as follows.
Consider row \(i\) of the matrix. We construct a Gershgorin disk for row \(i\) in the complex plane as follows. The disc is centered at the entry of the row that lies on the diagonal of the matrix, \(a_{ii}\). Its radius is given by the sum of the absolute values of all other entries of the row, \(\sum_{j\ne i} \left|a_{ij}\right|\).
Construct a Gershgorin disk for all rows of the matrix.
Every eigenvalue of \(\mathsf{A}\) will lie within at least one of the Gershgorin discs.
As an example of the Gershgorin circle theorem, consider the matrix
\begin{align} \mathsf{M} = \begin{pmatrix} -2 & 0 & 5 \\ 2 & -6 & 0 \\ 2 & 4 & 4 \end{pmatrix} \end{align}
It has one real eigenvalue at about \(5.7\), and two complex eigevalues near \(-4.9 \pm 1.5i\). The Gershgorin discs are all centered on the real axis (since the diagonal entries are all real), and the radii of the discs for the respective rows are 5, 2, and 6. The plot below shows the eigenvalues as ×’s in the complex plane along with the respective Gershgorin discs colored in blue, orange, and green.
[2]:
def gershgorin_plot(M, n_points=400, palette=colorcet.b_glasbey_category10):
"""Plot Gershgorin discs with eigenvalues"""
# Centers and radii of Gershgorin discs
center = np.diag(M)
radius = np.sum(np.abs(M), axis=1) - np.abs(center)
# Maximum extent of a disc
maxx = np.max(np.abs(center.real + np.sign(center.real) * radius))
maxy = np.max(np.abs(center.imag + np.sign(center.imag) * radius))
max_extent = max(maxx, maxy)
# Buffer extent
max_extent *= 1.05
# Set up figure
p = bokeh.plotting.figure(
frame_width=300,
frame_height=300,
x_axis_label="Re",
y_axis_label="Im",
x_range=[-max_extent, max_extent],
y_range=[-max_extent, max_extent],
)
# Fix axis locations
p.xaxis.fixed_location = 0
p.yaxis.fixed_location = 0
# No grid lines
p.xgrid.visible = False
p.ygrid.visible = False
# Plot the discs
t = np.linspace(0, 2 * np.pi, n_points)
for c, r, color in zip(center, radius, palette):
x = c + r * np.cos(t)
y = r * np.sin(t)
p.patch(x=x, y=y, fill_color=color, alpha=0.3, line_color=None)
# Compute and plot eigenvalues
lam = np.linalg.eigvals(M)
p.x(lam.real, lam.imag, color="black", size=10, line_width=2)
return p
M = np.array(
[
[-2, 0, 5],
[ 2, -6, 0],
[ 2, 4, 4],
]
)
bokeh.io.show(gershgorin_plot(M))
Application of the Gershgorin theorem to the delay circuit
Considering again the matrix \(\mathsf{A}\), we look at the last row. The diagonal entry is \(-\gamma\), so the Gershgorin disc is centered at \(-\gamma\) on the real axis.. There is only one off-diagonal entry, which has magnitude \(\gamma\), so the Gershgorin disc has radius \(\gamma\). Because it is centered at \(-\gamma\) and has radius \(\gamma\), the Gershgorin disc is entirely to the left of the imaginary axis, just touching it at the origin of the complex plane. This means that the maximum possible value for the real part of an eigenvalue lying in that Gershgorin disc is zero.
The same argument holds for rows 2 through \(N+1\). The Gershgorin discs are all centered at \(-\kappa\) on the real axis and have radius \(\kappa\).
The interesting row, then, is the first row. The Gershgorin disc is centered at \(-1\) on the real axis and its radius is \(b = n m_0^n/(1+m_0^n)\). The value of \(b\) is always less than one for \(n \le 1\). Thus, in order for any eigenvalue to have a positive real part, and therefore for the fixed point of the circuit to be linearly unstable, we must have \(n > 1\), meaning that ultrasensitive repression is a requirement for an oscillatory instability. The Gershgorin circle theorem allowed us to conveniently derive this requirement without computing eigenvalues directly.
Any instability must be oscillatory
Having established that ultrasensitive repression is necessary for an instability, we now will show that any instability is oscillatory. That is, if there is an eigenvalue with a positive real part, it is complex.
To demonstrate this, we write the characteristic polynomial of the linear stability matrix. It can be computed by taking the determinant of \(\mathsf{A} - \lambda \mathsf{I}\) using cofactors.
\begin{align} (1+\lambda)(\kappa + \lambda)^N(\gamma + \lambda) + b\kappa^N \gamma = 0. \end{align}
This is an \((N+2)\)-order polynomial in \(\lambda\). Without expanding the polynomial, it is evident that every term in the polynomial is positive. Therefore, by the Descartes sign rule, there are no positive real roots. So, if there is a root with a positive real part, it must be complex.
Linear stability of the delay circuit
While the above analytical results are useful, we still need to use a brute-force approach to compute the linear stability diagram. First, we code up functions to compute the fixed point, the linear stability matrix, and eigenvalues to determine stability.
[3]:
def fixed_point(beta, n):
"""Computes fixed point for the m⟶r...⟶p autorepression system.
"""
return scipy.optimize.brentq(
lambda x: x * (1 + x ** n) - beta,
0,
beta,
)
def lin_stab_matrix(beta, kappa, gamma, n, N):
"""Compute linear stability matrix for delay circuit."""
# Compute fixed point
m_0 = fixed_point(beta, n)
# b is the upper right entry in lin stab matrix
b = n * m_0 ** n / (1 + m_0 ** n)
# Construct linear stability matrix
A = kappa * (np.diag(-np.ones(N+2)) + np.diag(np.ones(N+1), -1))
A[0, 0] = -1.0
A[0, -1] = -b
A[-1, -2] = gamma
A[-1, -1] = -gamma
return A
def lin_stab(beta, kappa, gamma, n, N):
"""Returns True if delay system is linearly stable and False
otherwise.
"""
# Linear stability matrix
A = lin_stab_matrix(beta, kappa, gamma, n, N)
# Compute eigenvalues
lam = np.linalg.eigvals(A)
# Return True is all eigenvalues have negative real parts
return (lam.real < 0).all()
Before we proceed to computing the linear stability diagram, we take a moment to emphasize that the Gershgorin circle theorem only established that ultrasensitivity is only a necessary condition for an instability, not sufficient. Furthermore, having a Gershgorin disc cross the imaginary axis is a necessary, but not sufficient condition for an instability. We demonstrate that now for an illustrative parameter set with \(\beta = 2\), \(\gamma = 3\), \(\kappa = 2\), \(n = 3\) and \(N = 5\).
[4]:
# Parameters illustrating Gershgorin circle theorem
beta = 2
gamma = 3
kappa = 2
n = 3
N = 5
A = lin_stab_matrix(beta, kappa, gamma, n, N)
bokeh.io.show(gershgorin_plot(A, palette=["#1f77b4"] + ["gray"] * N + ["orange"]))
The mid-sized gray circle is the overlay of \(N = 5\) Gershgorin discs. The orange circle is the Gershgorin disc from the bottom row of the matric. The blue disc is from the first row and clearly could encompase eigenvalues with positive real parts. However, the two eigenvalues that lie with in the blue disc (and only within the blue disc, which can be seen by zooming in) do not have positive real parts.
We now turn to performing linear stability analysis on the circuit. We have five(!) parameters to scan. We will make the plot in the \(\kappa\)-\(\beta\) plane. We will then allow for tuning of the ratio of decay rates of protein and mRNA \(\gamma\), the Hill coefficient \(n\), and the number of intermediates \(N\) via sliders. To make the graphic more responsive, we will pre-compute the linear stability matrices for all of the values of \(\gamma\), \(n\), and \(N\). This takes a while to compute, since we are considering 500 different sets of \((\gamma, n, N)\) values with 10,000 sets of \((\beta, \kappa)\) values for a total of 5 million eigenvalue calculations.
[5]:
# Parameter values to compute stability
log_gamma_vals = np.arange(-2, 3)
n_vals = np.arange(1, 11)
N_vals = np.arange(1, 11)
beta_vals = np.logspace(-1, 4, 100)
kappa_vals = np.logspace(-3, 3, 100)
# Compute the linear stability diagram for various n and N and store in dict
stab_diagrams = {}
for n, N, log_gamma in tqdm.tqdm(itertools.product(n_vals, N_vals, log_gamma_vals)):
stab = np.empty((len(beta_vals), len(kappa_vals)))
gamma = 10.0**log_gamma
for i, beta in enumerate(beta_vals):
for j, kappa in enumerate(kappa_vals):
stab[i, j] = lin_stab(beta, gamma, kappa, n, N)
stab_diagrams[f"{n},{N},{log_gamma}"] = stab
500it [07:27, 1.12it/s]
Finally, we can make a figure. We will use the precomputed “images” for the stability in JavaScript callbacks for smooth interactivity. Again, we will color the unstable region in dark gray.
[6]:
# Plot the result (shaded region has oscillatory instability)
p = bokeh.plotting.figure(
frame_height=250,
frame_width=250,
x_axis_label="κ",
y_axis_label="β",
x_axis_type="log",
y_axis_type="log",
x_range=bokeh.models.Range1d(kappa_vals.min(), kappa_vals.max()),
y_range=bokeh.models.Range1d(beta_vals.min(), beta_vals.max()),
)
# Build the sliders
n_slider = bokeh.models.Slider(title="n", value=5, start=1, end=10, step=1, width=150)
N_slider = bokeh.models.Slider(title="N", value=5, start=1, end=10, step=1, width=150)
log_gamma_slider = bokeh.models.Slider(
title="γ",
value=0,
start=-2,
end=2,
step=1,
width=150,
format=bokeh.models.CustomJSTickFormatter(
code="return Math.pow(10, tick).toFixed(2)"
),
)
# Plot an image with Bokeh; encapsulate 2D array as a list
source = bokeh.models.ColumnDataSource(
data=dict(
image=[
stab_diagrams[f"{n_slider.value},{N_slider.value},{log_gamma_slider.value}"]
]
)
)
p.image(
image="image",
source=source,
x=kappa_vals.min(),
y=beta_vals.min(),
dw=kappa_vals.max(),
dh=beta_vals.max(),
palette="Greys3",
alpha=0.5,
)
# JavaScript code for the callback
js_code = """
let key = String(n_slider.value) + ','
+ String(N_slider.value) + ','
+ String(log_gamma_slider.value);
source.data['image'] = [stab_diagrams[key]];
source.change.emit();
"""
callback = bokeh.models.CustomJS(
args=dict(
source=source,
stab_diagrams=stab_diagrams,
n_slider=n_slider,
N_slider=N_slider,
log_gamma_slider=log_gamma_slider,
),
code=js_code,
)
# Link the callbacks
n_slider.js_on_change("value", callback)
N_slider.js_on_change("value", callback)
log_gamma_slider.js_on_change("value", callback)
# Build and display the layout1
layout = bokeh.layouts.row(
p,
bokeh.models.Spacer(width=15),
bokeh.layouts.column(n_slider, N_slider, log_gamma_slider),
)
bokeh.io.show(layout)